Ants, Bees, Genomes & Evolution @ Queen Mary University London
Published: 12 September 2011
Relaxed selection is a precursor to the evolution of phenotypic plasticity
B. Hunt, L. Ometto, Y. Wurm, D. Shoemaker, S. Yi, L. Keller, M. Goodisman
Proceedings of the National Academy of Sciences of the USA, 2011, 108:15936-41
Abstract
Phenotypic plasticity allows organisms to produce alternative phenotypes under different conditions and represents one of the most important ways by which organisms adaptively respond to the environment. However, the relationship between phenotypic plasticity and molecular evolution remains poorly understood. We addressed this issue by investigating the evolution of genes associated with phenotypically plastic castes, sexes, and developmental stages of the fire ant Solenopsis invicta. We first determined if genes associated with phenotypic plasticity in S. invicta evolved at a rapid rate, as predicted under theoretical models. We found that genes differentially expressed between S. invicta castes, sexes, and developmental stages all exhibited elevated rates of evolution compared with ubiquitously expressed genes. We next investigated the evolutionary history of genes associated with the production of castes. Surprisingly, we found that orthologs of caste-biased genes in S. invicta and the social bee Apis mellifera evolved rapidly in lineages without castes. Thus, in contrast to some theoretical predictions, our results suggest that rapid rates of molecular evolution may not arise primarily as a consequence of phenotypic plasticity. Instead, genes evolving under relaxed purifying selection may more readily adopt new forms of biased expression during the evolution of alternate phenotypes. These results suggest that relaxed selective constraint on protein-coding genes is an important and underappreciated element in the evolutionary origin of phenotypic plasticity.
Phenotypic plasticity is a fundamentally important process that allows organisms to develop distinct adaptive phenotypes in response to spatial and temporal variation in the environment (1). The most dramatic examples of phenotypic plasticity are embodied by polyphenisms, where multiple discrete phenotypic classes develop under different environmental conditions (2, 3). Examples of polyphenisms are particularly prominent in insects, which include beetle horns (4), aphid wing morphs (5), and social insect castes (6, 7). The specialized caste polyphenisms of social insect taxa, in particular, have been integral to their striking ecological success (6). Phenotypic plasticity thus plays an important role in the shaping of ecosystems and is a potential driver of evolutionary change (1, 8).
At a molecular level, alternate phenotypes produced through phenotypic plasticity develop through the differential expression of genes among individuals (9–15). Although differential gene expression may arise as an indirect consequence of phenotypic differentiation, many differentially expressed genes exhibit morph-biased fitness consequences (14) and signs of adaptive changes in gene expression (14, 15). As a result, large-scale investigations of gene expression differences among individuals with distinct phenotypes can provide insight into the molecular evolutionary basis of phenotypic differentiation (9–15).
Despite the broad importance of differential gene expression to phenotypic plasticity, the processes affecting the evolution of differentially expressed genes remain poorly understood. Elevated rates of protein evolution have been predicted for genes with differences in expression associated with distinct morphs (10, 16–19). However, few studies have investigated the molecular evolution of differentially expressed genes associated with phenotypic plasticity in cases where there are no fixed genetic differences among morphs (20, 21). Thus, an important and largely unanswered question is whether genes differentially expressed among alternate phenotypes are generally subject to elevated rates of evolution.
In addition, it is unclear if changes in the rate of molecular evolution result from (16, 17, 22, 23) or facilitate (19, 24) gene expression bias. Gene expression bias could lead to changes in rates of molecular evolution if selection becomes less effective at removing deleterious alleles when only a subset of the population expresses the focal gene (16, 17). Consequently, a relaxation of purifying selection on protein-coding genes could be a direct result of gene expression bias. Alternatively, the degree of purifying selection acting on a gene may be correlated with its propensity to deviate in expression pattern and become differentially expressed among alternate morphs during the course of evolution (19, 24–26). In particular, fast-evolving genes, which generally experience low levels of purifying selection, could more readily adopt biased expression patterns over time. Thus, genes differentially expressed among phenotypic classes could be fast-evolving either because of the way selection acts on differentially expressed genes or because fast-evolving genes have a propensity to become expressed differentially among alternate morphs over time.
We used the fire ant Solenopsis invicta and the honey bee Apis mellifera to investigate the molecular evolution of genes showing biased expression among alternate phenotypes. Hymenopteran social insects are ideal models for studying the link between gene expression and phenotypic diversity because they display remarkable examples of phenotypic plasticity in association with the reproductive division of labor characterizing their colonies (7). This is exemplified by the development of female queen and worker castes, where queens reproduce and workers engage in tasks related to brood-rearing and colony defense (7). Importantly, queens and workers exhibit extreme phenotypic differences but are not genetically differentiable in most taxa (6, 9, 27).
We further used S. invicta to investigate the molecular evolution of genes with expression bias between sexes and developmental stages. Hymenopterans are particularly well suited for such investigations because sex is determined by differences in ploidy level [hymenopteran males are typically haploid and females are diploid (28)] rather than by sex chromosomes. Moreover, hymenopterans undergo complete metamorphosis and display dramatic phenotypic differences among developmental stages [as in other holometabolous insects (12)], hence providing an additional comparison of alternate developmental phenotypes. Our analyses of castes, sexes, and developmental stages in S. invicta facilitate a direct comparison of the molecular evolution of loci associated with diverse forms of biased gene expression.
Results and Discussion
Gene Expression Bias Represents a Key Factor Associated with the Rate of Protein Evolution.
Microarray analyses conducted on whole bodies of adult and pupal males, queens, and workers of the fire ant S. invicta (Fig. 1A) revealed that many genes were differentially expressed between age-matched female castes, sexes, and developmental stages (15) (Fig. 1B). We investigated if gene expression bias was associated with the rate of gene evolution by analyzing coding sequences from the ants S. invicta (29), Pogonomyrmex barbatus (30), and Linepithema humile (31).
Fig. 1
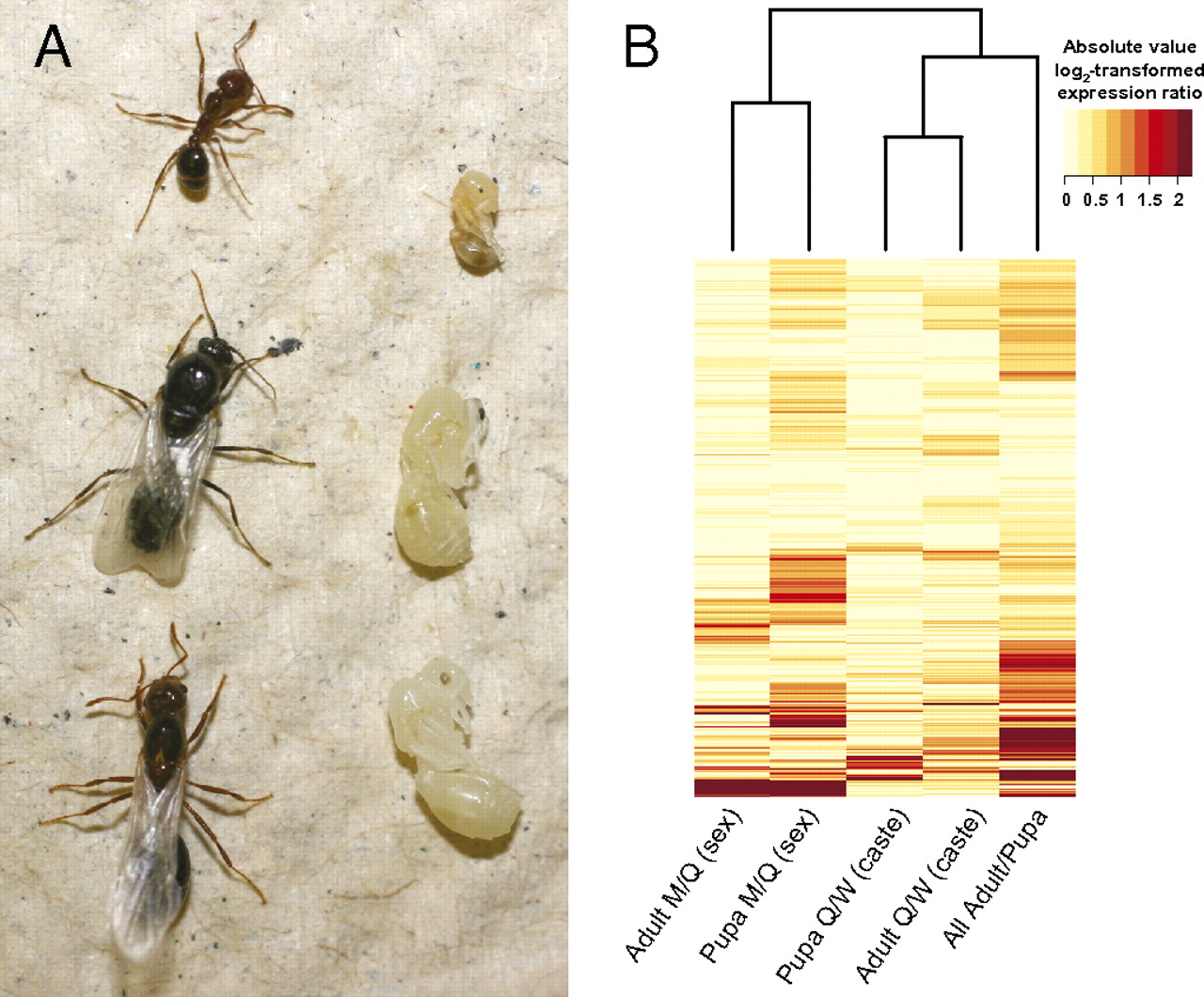
Distinct gene expression profiles associated with dramatic phenotypic differences in the fire ant S. invicta. (A) Adult (Left) and pupal (Right) worker, male, and queen (Top to Bottom). Photo credit: A. Palaski and A. Thompson. (B) Relationships among gene expression profiles of pupal and adult queens (Q), workers (W), and males (M) for 1,101 genes, where a log2-transformed expression ratio of 1 corresponds to a twofold difference in expression.
We found that the rate of protein evolution [measured as the ratio between the rate of nonsynonymous and synonymous substitutions (dN/dS)] in S. invicta was significantly correlated with two principal components representing the pooled effects of gene expression bias (PC1) and overall gene expression level (PC3) (32) (Fig. 2 and Fig. S1). The strengths of correlations between protein evolutionary rates and PC1 and PC3 were similar but in opposite directions (Fig. 2B). Thus, diverse forms of gene expression bias, when taken together, contribute to protein evolutionary rate variation at levels comparable to overall gene expression level in S. invicta. This demonstrates that gene expression bias among individuals and developmental stages is a key factor (32–34) associated with the rate of protein evolution.
Fig. 2
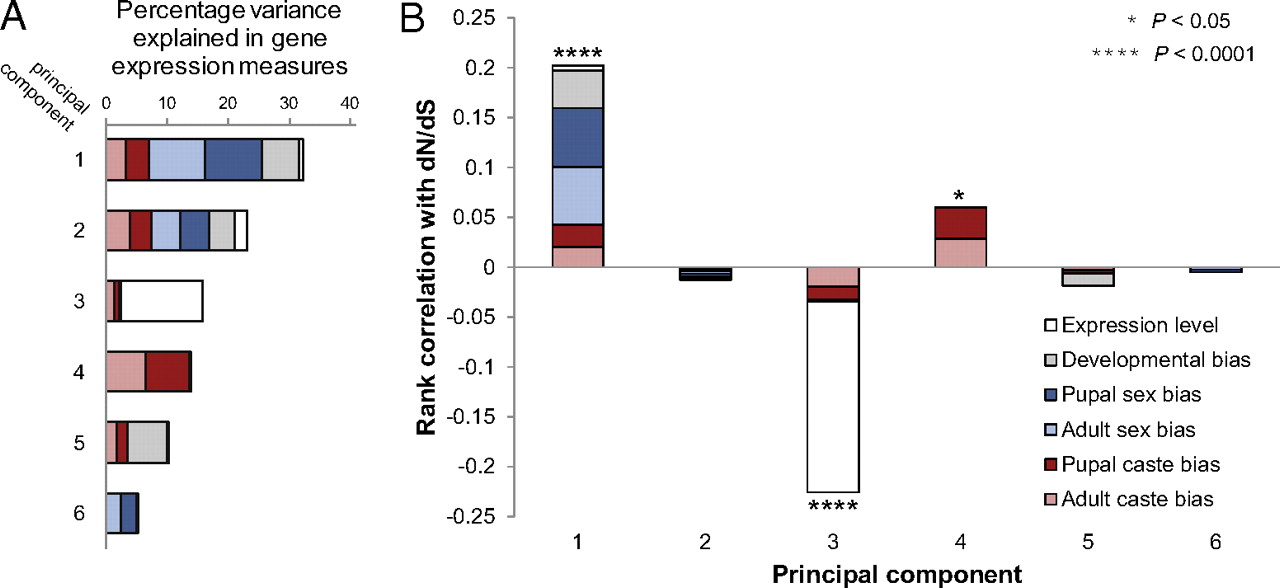
Associations between metrics of gene expression and protein evolution in the fire ant S. invicta. (A) Percentage variance explained by six principal components comprising expression level, developmental expression bias, pupal sex expression bias, adult sex expression bias, pupal caste expression bias, and adult caste expression bias. Colors depict the proportional contribution of each gene expression measure to each principal component. (B) Relative contribution of each gene expression measure to each principal component when normalized by the Spearman's rank correlation coefficient of each principal component with the S. invicta protein evolutionary rate dN/dS.
The correlations between the degree of gene expression bias and protein evolutionary rate were significant for all three instances of alternate phenotypes examined (Fig. S2). Furthermore, there was an approximately proportional relationship between the rate of protein evolution and degree of gene expression bias in all three cases, with the highest dN/dS value attributable to genes with greatest expression bias among morphs (Fig. 3A). The association between evolutionary rate and gene expression bias was also approximately additive; genes that showed bias in multiple different contexts (i.e., castes, sexes, developmental stages) tended to evolve more rapidly than genes that showed bias in only a single context (Fig. 3 B and C).
Fig. 3
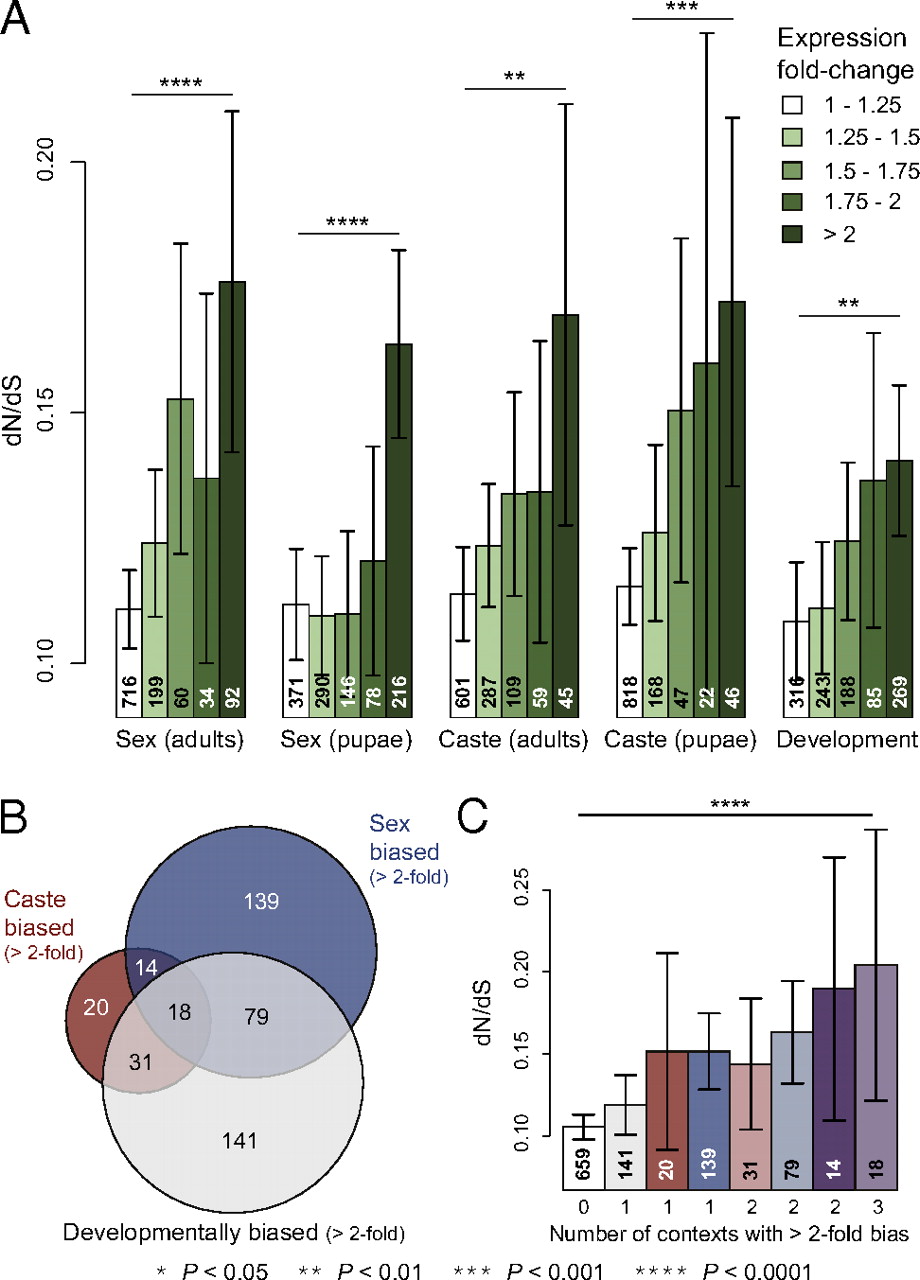
Relaxed selection is ubiquitously associated with gene expression bias in the fire ant S. invicta. (A) Protein evolutionary rate dN/dS for genes showing varying levels of differential expression between sexes, castes, and developmental stages. (B) Overlap of genes with greater than twofold difference in expression between sexes, castes, and developmental stages. (C) Protein evolutionary rate for each section of the Venn diagram in B compared with all genes exhibiting less than twofold expression difference between sexes, castes, and developmental stages. Means with 95% confidence intervals are plotted in A and C, the text in bars denotes the number of genes in each bin, and significance was determined by Kruskal–Wallis tests.
We conducted a separate analysis of the relationship between gene expression bias among castes and rate of protein evolution in the honey bee A. mellifera, which evolved sociality independent from ants. Previously, we analyzed gene expression data from A. mellifera queen and worker brain tissue (35) to highlight differences in evolution between queen- and worker-biased genes (20). In a reanalysis of these data, we found that, as in S. invicta, the overall degree of differential gene expression between A. mellifera queens and workers was positively associated with the rate of gene evolution (Fig. S3). This finding further supports the view that biased gene expression is associated with an increase in protein evolutionary rate in polyphenic social insects.
Purifying Selection Explains Most Variation in Protein Evolutionary Rate.
Variation in the rate of protein evolution may reflect differences in the strengths of positive selection or purifying selection. To understand the relative contributions of these different modes of selection better, we conducted several analyses to detect signatures of positive selection during S. invicta protein evolution.
These analyses suggested that positive selection was not dominant in shaping overall rates of protein evolution. Only 3 of 1,104 S. invicta genes with expression data and orthologs in other ant taxa exhibited clear evidence of positive selection when averaged across all codons [dN/dS > 1; for the remaining genes, mean dN/dS = 0.122 (SEM ± 0.003); Table S1]. Four additional genes (of a set of 365 as discussed in Methods) exhibited a robust signature of lineage-specific positive selection in S. invicta according to consensus results from branch-site tests (36) based on two sequence-alignment methods (37) (Methods and Table S2). Overall, the paucity of genes exhibiting strong signatures of positive selection suggests that the observed correlations between gene expression bias and protein evolutionary rates were shaped largely by variation in the strength of purifying selection (33, 38).
To explore this issue further, we compared sequence polymorphism within S. invicta with sequence divergence between the S. invicta lineage and the outgroup taxa P. barbatus and L. humile. There were 381 genes with identified SNPs in S. invicta, sequence divergence data among taxa, and gene expression data in S. invicta. McDonald–Kreitman tests (39) were used to investigate the type of selection operating on these 381 genes, partitioned according to different levels of expression bias.
We detected strongly significant (P = 3.4e-9) purifying selection in a pooled analysis of 223 genes showing low levels (i.e., less than 2-fold) of expression bias. There was also significant (P = 0.048) purifying selection detected in a pooled analysis of the 158 genes with greater than twofold expression bias in one or more contexts (Table S3). Thus, although it is likely that positive selection and demographic effects (40) contributed to some extent to the rate of molecular evolution, our data suggest that variation in protein evolutionary rate (Figs. 2 and 3) was largely driven by variation in the strength of purifying selection (33, 38). Moreover, the strength of purifying selection, as measured by the direction of selection (41), was stronger in genes not differentially expressed among morphs than in genes exhibiting differential expression (Table S3). This suggests that genes with biased expression evolved at elevated rates relative to unbiased genes, predominantly because purifying selection was weaker.
Rates of Protein Evolution and Different Phenotypic Forms.
The rate of evolution of both queen- and worker-biased genes in S. invicta adult and pupal stages was higher than the rate of evolution of unbiased genes. However, the difference was significant only for the queen-biased genes (20) (Figs. S4 and S5C). Theory predicts that genes expressed in workers but not in queens should experience a relaxation of purifying selection relative to genes expressed in queens but not in workers, because selection operates indirectly on the sterile worker caste (42). However, our results show that worker-biased genes did not evolve faster than queen-biased genes. Moreover, the rates of evolution for worker- and queen-biased genes did not differ substantially from those of sex- or developmentally biased genes (Fig. S4). There are at least two possible nonmutually exclusive explanations for these results. First, indirect selection acting on workers or colony-level traits in social insects may not be substantially weaker than selection acting on other types of conditional traits. Second, the lack of a higher rate of evolution of worker-biased genes may stem from the fact that none of these genes were exclusively expressed in workers (17, 42).
In the case of development, adult-biased genes disproportionately contributed to an increase in protein evolutionary rates relative to unbiased genes (although pupal- and adult-biased genes did not differ significantly from one another in evolutionary rate; Fig. S4). Furthermore, little difference was observed between the rates of evolution of male- and queen-biased genes in S. invicta. This stands in contrast to results from many studies of sex-biased gene expression, where male-biased genes typically have been the primary driver of increased evolutionary rates associated with sex (10, 43).
Fast-Evolving Genes Were Preferentially Recruited in the Evolution of Social Insect Castes.
We investigated whether the elevated rate of protein evolution observed for differentially expressed genes occurred before (19, 24) or after (17, 22, 23) the evolution of gene expression bias associated with phenotypic plasticity. To address this issue, we examined the evolution of proteins following the independent evolution of castes in ants and bees and in a parallel nonsocial lineage. We first identified 3,705 three-way orthologs of S. invicta, A. mellifera, and the nonsocial jewel wasp Nasonia vitripennis. In total, 1,050 of these orthologs were also associated with gene expression data from S. invicta castes (15).
Remarkably, the rate of evolution of genes in the N. vitripennis lineage was strongly associated with expression bias of the S. invicta orthologs (Fig. 4B). This result was further supported by an analysis of 1,289 orthologs (of the 3,705) for which expression data from A. mellifera castes were available (35). As was the case in S. invicta, the degree of caste bias of genes in A. mellifera was significantly associated with the rate of evolution of orthologs in N. vitripennis (Fig. 4C). Overall, these results support the view that rapid rates of protein evolution preceded biased gene expression associated with the evolution of castes and that fast-evolving genes were therefore recruited into the process of caste differentiation.
Fig. 4
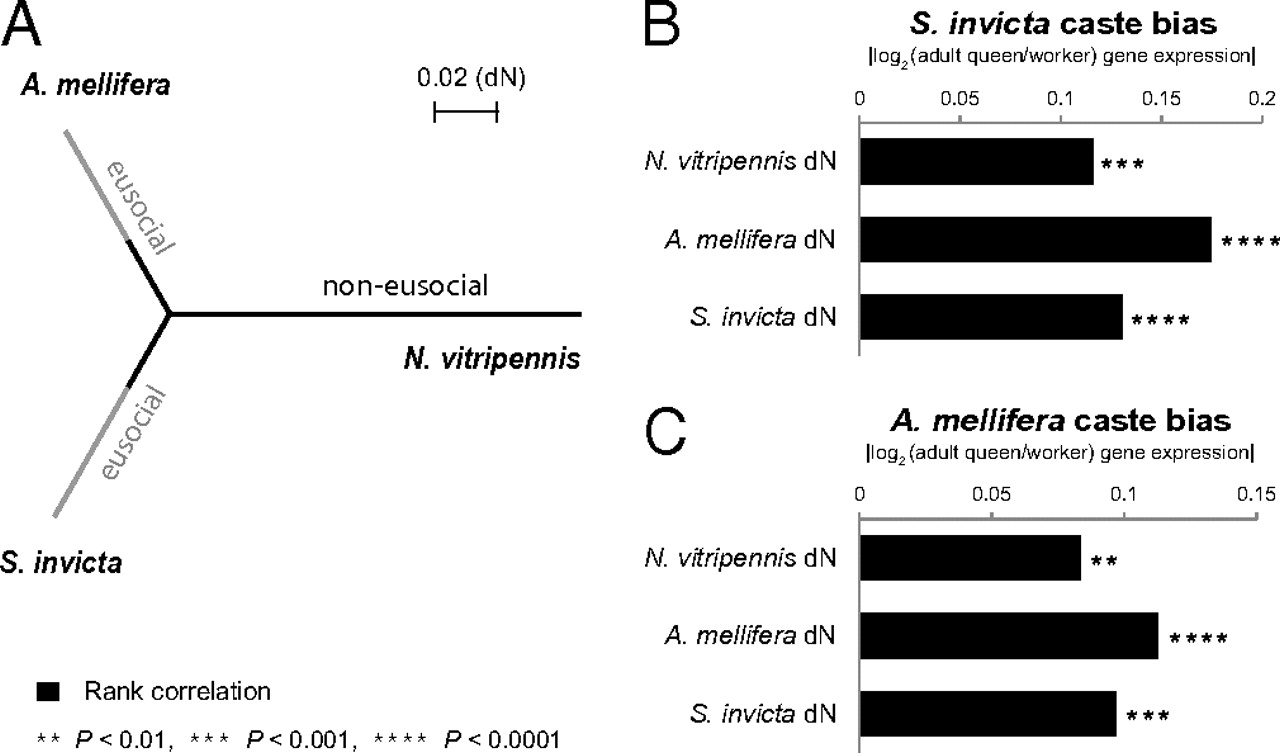
Evolutionary histories of genes with biased expression. (A) Phylogeny based on mean dN for 3,705 ortholog groups, including A. mellifera, S. invicta, and N. vitripennis. Gray lines denote the independent evolution of sociality in S. invicta and A. mellifera. (B) Spearman's rank correlations between the level of caste bias of genes (differential expression between queens and workers) in S. invicta and dN (n = 1,050 genes). (C) Spearman's rank correlations between the level of caste bias of genes in A. mellifera and dN (n = 1,289 genes).
The degree of differential expression of genes among castes was not significantly correlated between S. invicta and A. mellifera (Spearman’s ρ = 0.076, P = 0.136; n = 391). This lack of a significant correlation indicated that largely different sets of genes were differentially expressed between queens and workers in the two species, providing an additional opportunity to test whether fast-evolving genes were more likely to be recruited in the process of differential gene expression between castes. Our analyses revealed a strong association between the degree of differential expression in S. invicta and the rate of evolution of A. mellifera orthologs, and vice versa (Fig. 4B and C), as expected if relaxed selection preceded the evolution of caste systems in each of these two taxa.
Did Relaxed Selection Precede the Evolution of Biased Gene Expression in Social Insects?
Our results reveal that fast-evolving genes were preferentially recruited into caste-biased gene expression in both S. invicta and A. mellifera. This pattern could arise if genes under relaxed selective constraints were more likely to be coopted into mechanisms underlying the expression of alternate phenotypes. However, it is also possible that these genes already exhibited an elevated expression bias, among tissues (44) or sexes (10), for example, in the nonsocial ancestor of honey bees and ants. Indeed, a link between sex bias and tissue specificity has been observed in several taxa (24, 45), suggesting that tissue specificity and expression bias among morphs may be linked. If genes with bias in expression among tissues or between sexes are more likely to become differentially expressed in other contexts, social insect castes may have evolved largely by recruiting genes that were already involved in some type of phenotypic differentiation.
Unfortunately, the lack of data on gene expression in ancestors of A. mellifera and S. invicta makes it impossible to test whether tissue-specific or sex-biased genes have preferentially adopted caste-biased expression. However, it is possible to test whether the association between caste bias and sequence divergence is influenced by tissue specificity or sex bias in extant taxa. Such an analysis revealed that the observed correlations between caste-biased gene expression and protein evolutionary rate remained significant when controlling for either the degree of expression breadth in A. mellifera (46) or the sex- and developmentally biased gene expression in S. invicta (SI Text and Fig. S3). Although supporting the idea that tissue and sex specificity are not the main factors accounting for fast-evolving genes being preferentially adopted during the process of caste differentiation, the current analyses do not allow one to rule out the possibility that caste-biased genes displayed some other type of expression bias in ancestral populations, which could have contributed to the observed relaxation of purifying selection.
Concluding Remarks.
Our results suggest that genes with differential expression among alternate phenotypic forms evolve relatively rapidly because they are under relaxed selective constraint. Importantly, this association appears to stem primarily from fast-evolving genes being recruited in the process of phenotypic plasticity rather than from observed forms of gene expression bias themselves leading to relaxed purifying selection. Although relaxed selective constraint generally implies a diminished contribution to individual fitness at any given time point (e.g., 38), this lack of constraint may also facilitate specialized adaptation (18, 19, 47). In this manner, genes with limited selective constraints may play a particularly important role in the origin of phenotypic plasticity and the subsequent elaboration of specialized condition-specific phenotypes.
Methods
Gene Sequences.
The following official gene sets (OGSs) were used in our analyses: S. invicta OGS 2.2.0 (29), P. barbatus OGS 1.1 (30), L. humile OGS 1.1 (31), Harpegnathos saltator OGS 3.3 (48), A. mellifera OGS 1 (49), Nasonia vitripennis OGS 1.2 (50), and Drosophila melanogaster flybase release 5.21 (51). S. invicta, P. barbatus, L. humile, and H. saltator sequences are available from Fourmidable (52). A. mellifera, and N. vitripennis sequences are available from the Hymenoptera Genome Database (53).
Ortholog Determination.
Pairwise orthology between proteins from each species was assigned based on reciprocal BlastP (54) hits with an E value cutoff of E < 1e-10. BlastP output was then parsed with a custom BioPerl script (55). Pairwise reciprocal best hits and orthologous groups with three-way shared orthology and four-way shared orthology were assigned using custom Perl scripts. A total of 6,900 three-way orthologs between S. invicta, P. barbatus, and L. humile were found (5,598 with informative evolutionary rate data after filtering). A total of 4,839 three-way orthologs between S. invicta, A. mellifera, and N. vitripennis were found (3,705 with informative evolutionary rate data after filtering). A total of 5,476 four-way orthologs among the ant taxa were found (4,248 with informative evolutionary rate data after filtering).
Sequence Alignment.
MUSCLE 3.8.31 (56) was used to generate protein alignments for each orthologous group with default settings. PAL2NAL v13 (57) was then used to back-translate codon alignments from protein alignments and Gblocks 0.91b (58) was used to extract confidently aligned regions. To confirm branch-site tests of positive selection, a separate alignment procedure (37) was undertaken. In this case, PRANK (59) (release 100802) was used to generate codon alignments with default settings.
Evolutionary Rates.
PhyML 3.0 (60) was used to generate maximum likelihood phylogenies from codon alignments for each orthologous group with the “HKY85” nucleotide substitution model. PAML 4.4b (61) was then used to estimate dN and dS and ω (dN/dS) for each orthologous group using a phylogenetic tree produced by PhyML as input and the “F3 × 4” codon frequency model.
For three-way orthologs between S. invicta, P. barbatus, and L. humile and three-way orthologs between S. invicta, A. mellifera, and N. vitripennis, substitution rates were averaged across all codons for a given protein (NSsites = 0) with free dN/dS ratios for each branch (model = 1). Three genes with evidence of positive selection (dN/dS > 1) in S. invicta were identified utilizing this approach. In each case, dS was zero and dN/dS values were infinite. These three genes were eliminated from further analysis to avoid skewing means but are summarized in Table S1. In our three-way comparison of S. invicta, P. barbatus, and L. humile, the mean dS value for the S. invicta branch was 0.36 (SEM ± 0.006; maximum dS = 2.12), indicating that synonymous sites have not been saturated by multiple substitutions and analysis of dN/dS is appropriate. In our three-way comparison of S. invicta, A. mellifera, and N. vitripennis, the mean dS value for the S. invicta branch was 3.94 (SEM ± 0.191), indicating widespread synonymous site saturation. We therefore only used dN for comparing these more divergent taxa.
To test for further evidence of positive selection, we used the branch-site test of positive selection (branch-site model A, test 2) (36), which uses likelihood ratio tests to detect positive selection on a small number of sites along a specific lineage. For this test, we used MUSCLE alignments of four-way orthologs among the ants S. invicta, P. barbatus, L. humile, and H. saltator to construct phylogenetic trees using PhyML as before. S. invicta was set as the foreground branch, with two dN/dS ratios for branches (model = 2). For the alternative model, ω2 was estimated from the data (fix_omega = 0, omega = 1), whereas the null model fixes ω2 at 1 (fix_omega = 1, omega = 1) as described in PAML documentation and sample files (61). The log-likelihoods for the null and alternative models were used to calculate a likelihood ratio test statistic, which was then compared against the χ2df = 1 distribution (with a critical value of 3.84 at a 5% significance level) (61). P values were Bonferroni-corrected according to the number of tests of significance performed (n = 861; α/n = 5.8e-5 used as the threshold for significance). This highly conservative threshold was implemented because of the documented occurrence of false-positive results in branch-site tests of positive selection as a result of alignment errors (37). To control better for the influence of alignment errors, PRANK codon alignments were used as input in an alternative analysis of positive selection (37). For analysis of PRANK codon alignments, a Bonferroni cutoff of 1.4e-4 was used (n = 365). The consensus set of significant genes was then taken. Forty of 861 orthologs with MUSCLE alignments in our dataset retained a significant signal of positive selection on the S. invicta branch, whereas 5 of 365 orthologs with PRANK alignments in our dataset retained a significant signal of positive selection on the S. invicta branch. The consensus set consisted of four proteins (Table S2).
In each analysis of evolutionary rates, only genes with a length of ≥100 codons analyzed by PAML were used in our results. For analyses of evolutionary rates using four-way orthologs, only ortholog groups exhibiting the most common phylogeny topology were used. Phylogeny topology, foreground branch specification, branch-specific substitution rates, and log-likelihoods were processed and parsed using custom Perl scripts.
Polymorphism Analysis.
Sequencing reads from normalized S. invicta cDNA libraries were accessed from the National Center for Biotechnology Information Sequence Read Archive, including those generated from 100 heads of workers and queens from the United States (accession nos. SRR060014 and SRR060015) and those generated from two males each from 12 colonies in Argentina (accession nos. SRR060008, SRR060009, and SRR060010). Reads were assembled to a subset of OGS coding sequences with the highest possible quality rank (discussed in section on S. invicta array probe to genome mapping) using SSAHA2 with only uniquely high-scoring matches with 98% identity accepted (62). SAMtools (63) was used to assess variant sites utilizing “mpileup” and “bcftools.” The SAMtools Perl script “vcfutils.pl” was then used to filter variants for an rms quality value of 20, a minimum read depth of 4, and a distance of at least 10 bases from a gap. Alternate coding sequences with alternate alleles substituted were then generated using a custom Perl script. We used PAML to calculate nonsynonymous and synonymous polymorphisms in S. invicta by utilizing “codeml” to compare alternate and reference sequences. Because few SNPs per locus were detected, we performed subsequent analyses on pooled categories of genes (rounding codeml pooled maximum likelihood substitutions to the nearest integer; Table S3). Fixed substitutions used for further analyses of neutrality (39, 41) were taken as the number of synonymous and non-synonymous substitutions on the S. invicta branch in the three-way comparison of ants described above.
S. invicta Gene Expression.
Gene expression measures were obtained using custom cDNA microarrays (64, 65). We estimated the relative expression of each clone for whole-body age-matched adult and pupal queens, workers, and males using the Bayesian approach implemented in the program Bayesian Analysis of Gene Expression Levels [BAGEL (66)] as described previously (15). This BAGEL expression level was used to determine all log2-transformed ratios of gene expression among phenotypic groups. We used the absolute value of log2-transformed pairwise BAGEL expression ratios to measure the degree of sex bias in expression (male vs. queen) and caste bias in expression (queen vs. worker) for adults and pupae separately. To measure developmental bias, the absolute value of the log2-transformed ratio of summed adult male, worker, and queen BAGEL values vs. summed pupal male, worker, and queen BAGEL values was taken.
The overall gene expression level for each of the array clones, which was not included in prior analysis, was calculated as follows. For each hybridization experiment, gene expression level was estimated for each clone as the ratio between its net signal intensity and the average net signal intensity across clones. In particular, for each hybridization experiment, h, we assigned to each clone, c, a relative expression
where n is the number of S. invicta good-quality clones (as flagged in GenePix, Axon Instruments, Foster City, CA) and F and B are, respectively, the foreground and background signal intensities for either the green or red channel, depending on the labeling of the sample. Then, the overall gene expression level for each clone was calculated across all k hybridizations as
S. invicta Array Probe to Genome Mapping.
Sanger EST sequences of array clones were mapped to S. invicta genome scaffolds (assembly Si_gnF) using GMAP version 2010-03-09 (67). The best hit for each EST sequence was taken according to the genomic coordinates with the highest percent identity, and only ESTs with ≥95% identity and 50% coverage were retained. Genome coordinates and strand information were extracted for each S. invicta gene from the OGS 2.2.0 general feature format (GFF) file, which was subsequently used to map ESTs to genes using a custom Perl script.
The mean relative BAGEL expression level and the overall gene expression level of duplicate spots on the array were first calculated, and the R aggregate function was then used to take the mean of expression values for all clones that mapped to a given gene. This procedure resulted in gene expression measures for 2,088 unique genes (represented by 4,624 array clones). We filtered our dataset to include only S. invicta genes with proteins represented by a single cDNA and the highest possible quality rank (as indicated by the tag “quality = 5”).
S. invicta Gene Characteristics.
Normalized CpG dinucleotide content acted as a metric of levels of DNA methylation and was calculated as described previously (68) (Table S4). Third-codon position synonymous guanine-cytosine (GC) content and the frequency of optimal codons (Fop) were calculated using CodonW (http://codonw.sourceforge.net). Fop was calculated with automatic detection implemented to use the top 5% genes (in terms of bias in codon use). Coding sequence length and intron counts were calculated from the OGS 2.2.0 GFF file using a custom Perl script.
A. mellifera Gene Expression.
Data on gene expression bias between brains of queens and workers in A. mellifera were obtained from a study by Grozinger et al. (35). Data on expression breadth among tissues in A. mellifera were obtained from a study by Foret et al. (46) and processed as described previously (69). RNA-Seq data on expression levels in A. mellifera whole-body workers were obtained from a study by Zemach et al. (70).
Supporting Information
Supporting Information – (.PDF, 778.75 KB)
References
-
MJ West-Eberhard Developmental Plasticity and Evolution (Oxford Univ Press, New York, 2003).
-
HF Nijhout, Development and evolution of adaptive polyphenisms. Evol Dev 5, 9–18 (2003).
-
NA Moran, The evolutionary maintenance of alternative phenotypes. Am Nat 139, 971–989 (1992).
-
AP Moczek, DJ Emlen, Proximate determination of male horn dimorphism in the beetle Onthophagus taurus (Coleoptera: Scarabaeidae). J Evol Biol 12, 27–37 (1999).
-
CB Müller, IS Williams, J Hardie, The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecol Entomol 26, 330–340 (2001).
-
B Hölldobler, EO Wilson The Ants (Belknap Press of Harvard Univ Press, Cambridge, MA, 1990).
-
EO Wilson The Insect Societies (Harvard University Press, Cambridge, MA, 1971).
-
DW Pfennig, et al., Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol Evol 25, 459–467 (2010).
-
CR Smith, AL Toth, AV Suarez, GE Robinson, Genetic and genomic analyses of the division of labour in insect societies. Nat Rev Genet 9, 735–748 (2008).
-
H Ellegren, J Parsch, The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet 8, 689–698 (2007).
-
JF Ayroles, et al., Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41, 299–307 (2009).
-
BR Graveley, et al., The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 (2011).
-
G Gibson, The environmental contribution to gene expression profiles. Nat Rev Genet 9, 575–581 (2008).
-
T Connallon, AG Clark, Association between sex-biased gene expression and mutations with sex-specific phenotypic consequences in Drosophila. Genome Biol Evol 3, 151–155 (2011).
-
L Ometto, D Shoemaker, KG Ross, L Keller, Evolution of gene expression in fire ants: The effects of developmental stage, caste, and species. Mol Biol Evol 28, 1381–1392 (2011).
-
EC Snell-Rood, JD Van Dyken, T Cruickshank, MJ Wade, AP Moczek, Toward a population genetic framework of developmental evolution: The costs, limits, and consequences of phenotypic plasticity. Bioessays 32, 71–81 (2010).
-
JD Van Dyken, MJ Wade, The genetic signature of conditional expression. Genetics 184, 557–570 (2010).
-
R Gadagkar, The evolution of caste polymorphism in social insects: Genetic release followed by diversifying evolution. J Genet 76, 167–179 (1997).
-
JE Mank, H Ellegren, Are sex-biased genes more dispensable? Biol Lett 5, 409–412 (2009).
-
BG Hunt, et al., Sociality is linked to rates of protein evolution in a highly social insect. Mol Biol Evol 27, 497–500 (2010).
-
EC Snell-Rood, et al., Developmental decoupling of alternative phenotypes: Insights from the transcriptomes of horn-polyphenic beetles. Evolution 65, 231–245 (2011).
-
JA Brisson, SV Nuzhdin, Rarity of males in pea aphids results in mutational decay. Science 319, 58 (2008).
-
MS Barker, JP Demuth, MJ Wade, Maternal expression relaxes constraint on innovation of the anterior determinant, bicoid. PLoS Genet 1, e57 (2005).
-
JE Mank, L Hultin-Rosenberg, M Zwahlen, H Ellegren, Pleiotropic constraint hampers the resolution of sexual antagonism in vertebrate gene expression. Am Nat 171, 35–43 (2008).
-
JM Good, CA Hayden, TJ Wheeler, Adaptive protein evolution and regulatory divergence in Drosophila. Mol Biol Evol 23, 1101–1103 (2006).
-
P Khaitovich, et al., Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309, 1850–1854 (2005).
-
T Schwander, N Lo, M Beekman, BP Oldroyd, L Keller, Nature versus nurture in social insect caste differentiation. Trends Ecol Evol 25, 275–282 (2010).
-
GE Heimpel, JG de Boer, Sex determination in the Hymenoptera. Annu Rev Entomol 53, 209–230 (2008).
-
Y Wurm, et al., The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci USA 108, 5679–5684 (2011).
-
CR Smith, et al., Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci USA 108, 5667–5672 (2011).
-
CD Smith, et al., Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci USA 108, 5673–5678 (2011).
-
DA Drummond, CO Wilke, Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 (2008).
-
DP Wall, et al., Functional genomic analysis of the rates of protein evolution. Proc Natl Acad Sci USA 102, 5483–5488 (2005).
-
C Pál, B Papp, MJ Lercher, An integrated view of protein evolution. Nat Rev Genet 7, 337–348 (2006).
-
CM Grozinger, YL Fan, SER Hoover, ML Winston, Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera). Mol Ecol 16, 4837–4848 (2007).
-
J Zhang, R Nielsen, Z Yang, Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22, 2472–2479 (2005).
-
W Fletcher, Z Yang, The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol 27, 2257–2267 (2010).
-
AE Hirsh, HB Fraser, Protein dispensability and rate of evolution. Nature 411, 1046–1049 (2001).
-
JH McDonald, M Kreitman, Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991).
-
J Parsch, Z Zhang, JF Baines, The influence of demography and weak selection on the McDonald-Kreitman test: An empirical study in Drosophila. Mol Biol Evol 26, 691–698 (2009).
-
N Stoletzki, A Eyre-Walker, Estimation of the neutrality index. Mol Biol Evol 28, 63–70 (2011).
-
TA Linksvayer, MJ Wade, Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 63, 1685–1696 (2009).
-
Y Zhang, D Sturgill, M Parisi, S Kumar, B Oliver, Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450, 233–237 (2007).
-
L Duret, D Mouchiroud, Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol Biol Evol 17, 68–74 (2000).
-
RP Meisel, Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein-coding sequence evolution. Mol Biol Evol 28, 1893–1900 (2011).
-
S Foret, R Kucharski, Y Pittelkow, GA Lockett, R Maleszka, Epigenetic regulation of the honey bee transcriptome: Unravelling the nature of methylated genes. BMC Genomics 10, 472 (2009).
-
R Bonduriansky, SF Chenoweth, Intralocus sexual conflict. Trends Ecol Evol 24, 280–288 (2009).
-
R Bonasio, et al., Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068–1071 (2010).
-
Honeybee Genome Sequencing Consortium, Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949 (2006).
-
JH Werren, et al., Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science; Nasonia Genome Working Group 327, 343–348 (2010).
-
MA Crosby, JL Goodman, VB Strelets, P Zhang, WM Gelbart, FlyBase: Genomes by the dozen. Nucleic Acids Res; FlyBase Consortium 35, D486–D491 (2007).
-
Y Wurm, et al., Fourmidable: A database for ant genomics. BMC Genomics 10, 5 (2009).
-
MC Munoz-Torres, et al., Hymenoptera Genome Database: Integrated community resources for insect species of the order Hymenoptera. Nucleic Acids Res 39, D658–D662 (2011).
-
SF Altschul, et al., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997).
-
JE Stajich, et al., The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12, 1611–1618 (2002).
-
RC Edgar, MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004).
-
M Suyama, D Torrents, P Bork, PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609–W612 (2006).
-
G Talavera, J Castresana, Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56, 564–577 (2007).
-
A Löytynoja, N Goldman, An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA 102, 10557–10562 (2005).
-
S Guindon, O Gascuel, A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704 (2003).
-
ZH Yang, PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591 (2007).
-
Z Ning, AJ Cox, JC Mullikin, SSAHA: A fast search method for large DNA databases. Genome Res 11, 1725–1729 (2001).
-
H Li, et al., The Sequence Alignment/Map format and SAMtools. Bioinformatics; 1000 Genome Project Data Processing Subgroup 25, 2078–2079 (2009).
-
Y Wurm, J Wang, L Keller, Changes in reproductive roles are associated with changes in gene expression in fire ant queens. Mol Ecol 19, 1200–1211 (2010).
-
J Wang, et al., An annotated cDNA library and microarray for large-scale gene-expression studies in the ant Solenopsis invicta. Genome Biol 8, R9 (2007).
-
J Townsend, D Hartl, Bayesian analysis of gene expression levels: Statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol 3, RESEARCH0071–RESEARCH0071.0016 (2002).
-
D Wu, CK Watanabe, GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
-
N Elango, BG Hunt, MAD Goodisman, SV Yi, DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci USA 106, 11206–11211 (2009).
-
BG Hunt, JA Brisson, SV Yi, MAD Goodisman, Functional conservation of DNA methylation in the pea aphid and the honeybee. Genome Biol Evol 2, 719–728 (2010).
-
A Zemach, IE McDaniel, P Silva, D Zilberman, Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010).