Ants, Bees, Genomes & Evolution @ Queen Mary University London
Major bioinformatic software tools that we develop include:
SequenceServer - implement, visualize & interpret BLAST analyses
- BLAST is the most commonly used bioinformatics tool. But setting it up for private data and using it is counter-intuitive. We developed SequenceServer to make BLAST easy to use. Used in hundreds of laboratories worldwide.
- Sequenceserver: a modern graphical user interface for custom BLAST databases. Mol Biol Evol (2019).

CompareGenomeQualities - figure out which genome assembly is the best
- CompareGenomeQualities ranks assemblies based on contiguity, completeness, and accuracy. Contiguity is assessed through NG50 metric. Completeness and accuracy are assessed in genic regions using BUSCO and genome-wide using short Illumina reads. The tool is flexible, easy to run, and provides useful output visualisation.
- Parameter exploration improves the accuracy of long-read genome assembly.


Flo - transfer existing gene predictions to a new genome assembly
- Flo enables you to 'lift over' the gene prediction and other annotations from a GFF file to a new genome assembly. This is similar to UCSC LiftOver and NCBI LiftUp, but works with unpublished data on any species. It is multithreaded to ensure that it runs fast.

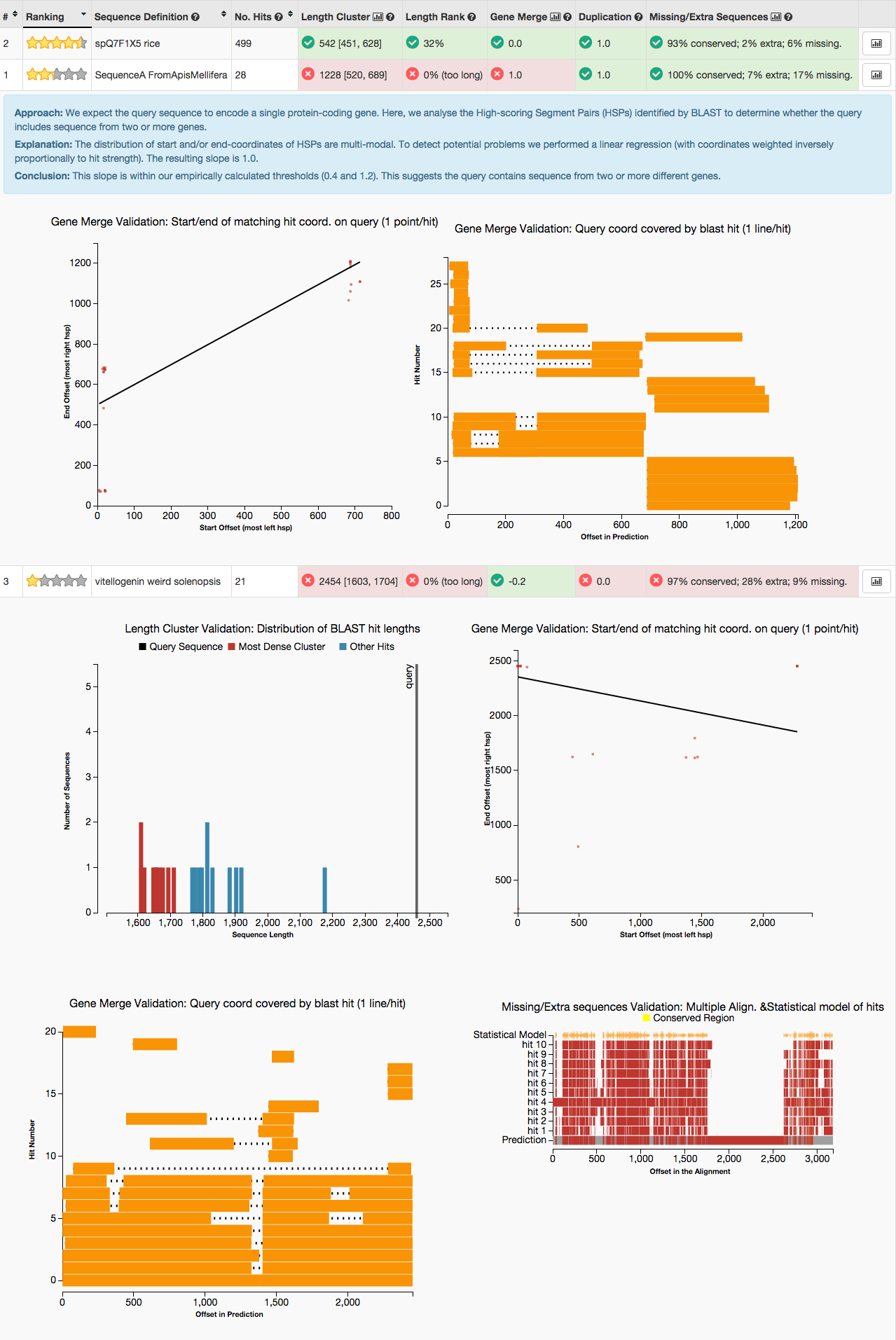
GeneValidator - choose the best gene models
- Tool to identify problematic gene predictions based on comparison with gene predictions in public databases. Funded by NESCent Google Summer of Code 2013. Useful for manual annotation, and for automatically choosing between multiple gene predictions for a given locus.
- GeneValidator: identify problems with protein-coding gene predictions. Bioinformatics (2016).
- https://github.com/wurmlab/genevalidator

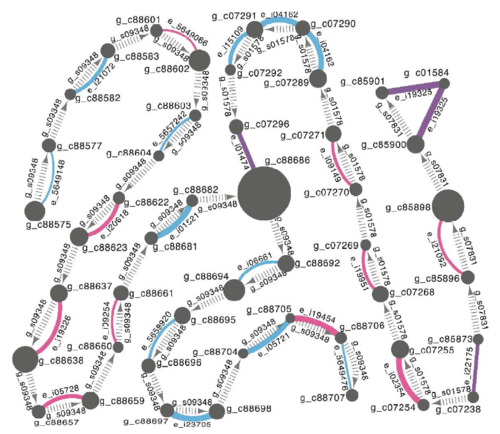
TGNet - evalutate genome quality with an independent transcriptome
- TGNet is a Cytoscape-based tool for visualization and quality assessment of de novo genome assemblies. Specifically it facilitates rapid detection of inconsistencies between a genome assembly and an independently derived transcriptome assembly. Developed by Oksana Riba-Grognuz. Used for fire ant genome project.
- Visualization and quality assessment of de novo genome assemblies. Bioinformatics (2011).
- https://github.com/ksanao/TGNet

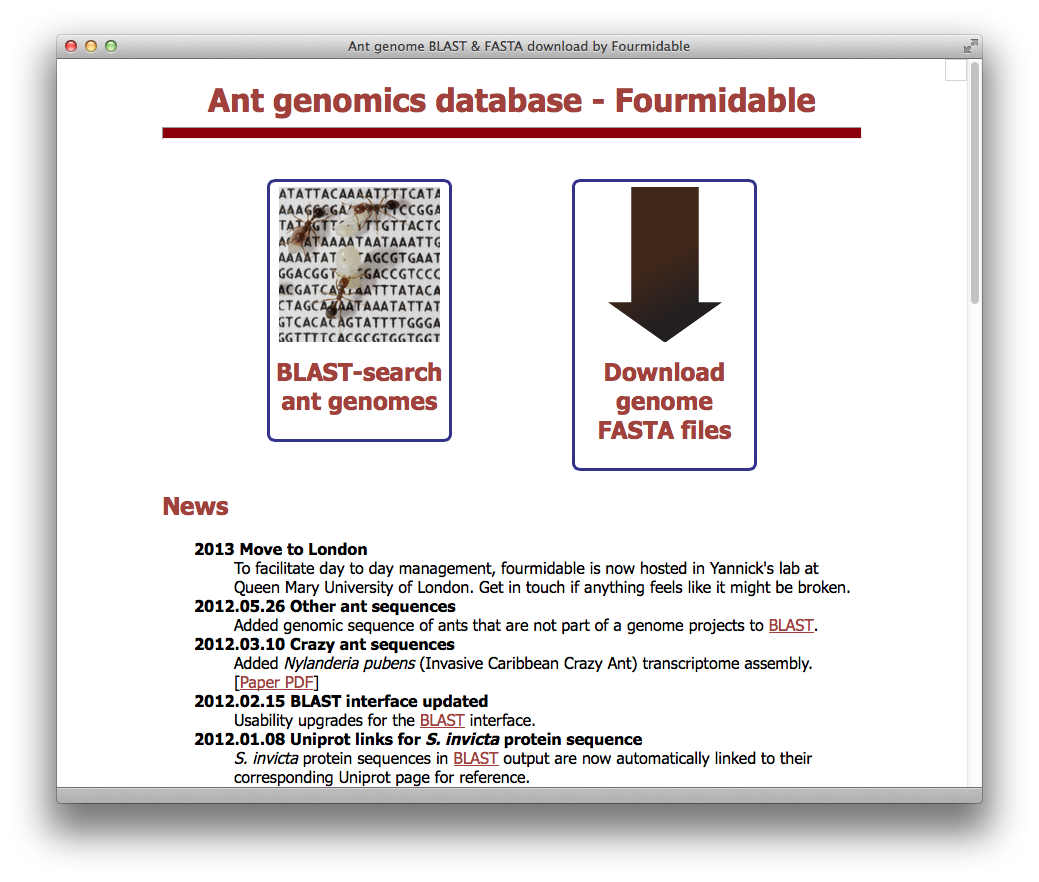
Fourmidable / antgenomes.org
- Community database centralizing genomic and transcriptomic sequence information for ants and related species (e.g., bees and wasps). Provides BLAST search and data download. Updated periodically with latest data and new tools.
- Fourmidable: a database for ant genomics. BMC Genomics 10:5 (2009).

Software development approaches
We believe in Reproducible Research & Software Sustainability. When possible we respect style guides and use unit testing & continuous integration. We aim to respect these approaches in our research on specific biological questions and in our tool development. We develop mostly in:
- R for number crunching.
- Shell for minimalisting pipelining.
- Python or ruby when text needs to be analysed.
- javascript/html/css for client-side stuff and some bioinformatics analysis pipelines
- But also use bash/python/perl when convenient or necessary...
But we love quick and dirty hacks too .
A lot of our code is at https://github.com/wurmlab.
Some tools now have limited relevance
oSwitch has been superceded by updates in docker. Afra has been used in our MSc course; it is currently somewhat out of sync with main Apollo.

BioNode
- BioNode is Node.js JavaScript library for client and server side bioinformatics.
- https://bionode.io


oswitch
- Provides quick and simple Docker based virtual environments on Macs and compute clusters alike for reproducible bioinformatics. Funded by NERC Environmental 'omics (EOS) Cloud. Given improvements to Docker this isn't really needed anymore.
- https://github.com/wurmlab/oswitch

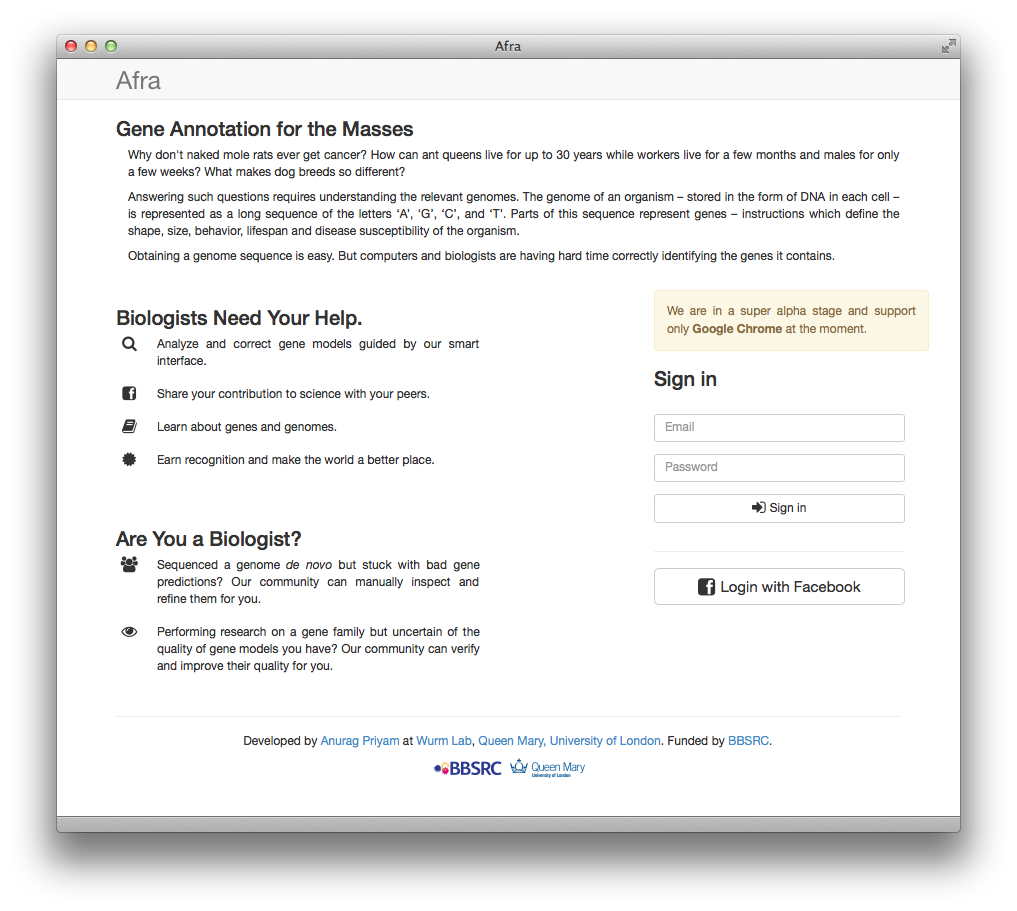
Afra
- BBSRC-funded tool for crowd-sourcing curation of gene predictions.
- https://github.com/wurmlab/afra
- Beta. Used in Bioinforatics MSc course at QMUL.
