Ants, Bees, Genomes & Evolution @ Queen Mary University London
UNIX/bioinformatics teaching cloud
Getting into big data science can be a big leap if you’re a biologist who is new to the command-line.
We try to cut that down into a series of smaller, more manageable steps.
As part of that, we run a hands-on genome bioinformatics course that introduces students to UNIX, and covers topics from Illumina read cleaning to genome assembly, annotation, population genomics and genome-wide association mapping.
For obvious 2020 reasons, we needed to do this online in a manner that:
- has manageable costs but sufficient power for genomics analyses;
- is easy for students to access autonomously;
- provides students the flexibility to work when they want (timezones) from where they want;
- can be easily modified by us as needed.
- doesn’t require students to install complex software (Docker, Virtual box, linux subsystem…) which are difficult to troubleshoot.
We built it.
Process for students
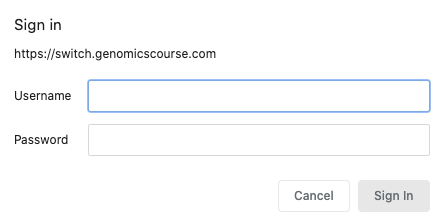
A student wanting to access their Linux machine must:
- connect to http://switch.genomicscourse.com (currently offline)
- enter their login/password
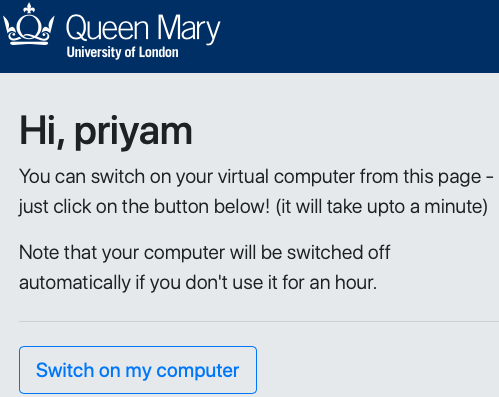
- click a button to switch on their virtual computer
- this creates their personal virtual computer and shows its IP address & hostname
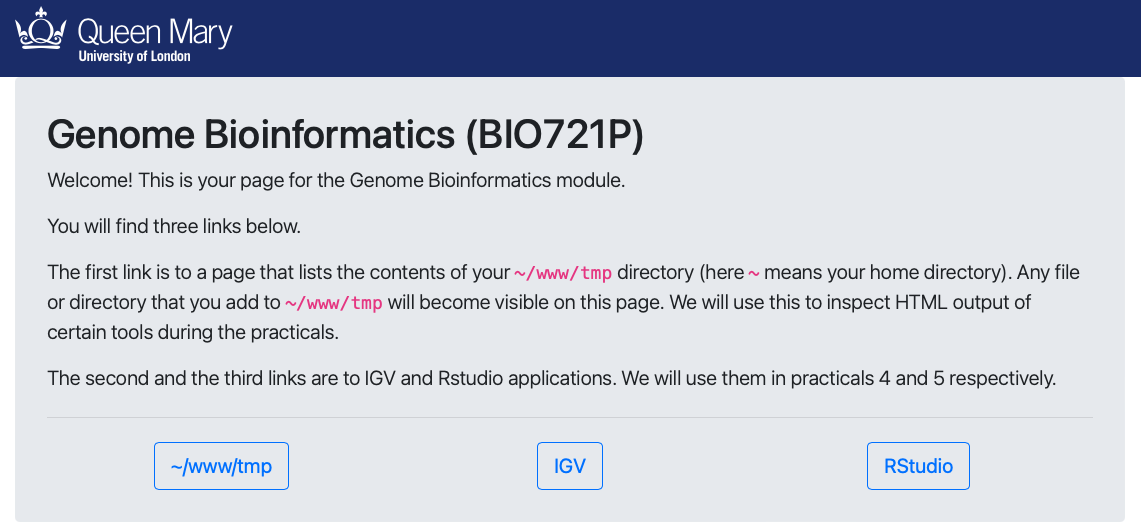
The student can connect to the computer by ssh, and download or visualise files in a web browser by putting them in a designated folder in their home directory (~/www).
If a student forgets to switch off their computer, this occurs automatically after 30 minutes of idle time.
For course administrators
This is great for us as organisers because:
- it avoids paying for cloud computing instances that are not being used.
- it allows us to give students more CPU and RAM.
- no physical rooms required - anyone can connect from any computer.
- ensures that all students use the same setup.
This uses Amazon EC2 infrastructure, and thus scales easily to any number of students, and can use computers with small or large amount of cpu power or ram.
Interested?
We can potentially deploy our solution for other courses. If you’re interested, get in touch.
Example screenshots
November 1, 2020